An investigation of two closely related proteins from a pathogenic bacterium has illustrated for the first time how evolution can shape the use of essential metals by enzymes. The research, conducted by an international team led by Professors Thomas Kehl-Fie (MMG) from the Department of Microbiology and Kevin Waldron from Newcastle University in the UK, was published in Nature Communications.
Almost half of all enzymes need an essential metal cofactor for catalysis, termed metalloenzymes. The abundance of metalloenzymes makes understanding the principles that govern metal-protein interactions relevant to nearly all aspects of biology, medicine, and biotechnology.
Metalloenzymes are often highly specific for their cognate metal ion cofactor, exhibiting reduced catalytic activity when bound to the wrong metal in vitro and in vivo. However, the features that dictate which metal is utilized by metalloenzymes are poorly understood. This limits our ability to manipulate metalloenzymes to produce novel synthetic enzymes that could perform useful chemical reactions for biotechnological applications or to develop metalloenzyme inhibitors for industrial and medical applications, including as antimicrobial drugs. The ubiquitous iron/manganese superoxide dismutase (SOD) family exemplifies this deficit in knowledge, as the specific metal used by any family member cannot be predicted in silico.
“Our work has broad implications for understanding how enzymes use essential metals for catalysis, and how this use of metals changes over evolutionary time,” Waldron said.
The group of researchers from the University of Illinois at Urbana-Champaign, Newcastle University, and Université Paris-Saclay in France, had previously demonstrated that an unusual pair of SOD metalloenzymes in the pathogenic bacterium Staphylococcus aureus, including methicillin resistant S. aureus (MRSA), play an important role during infection. They found that these SOD metalloenzymes differentially defend the bacterium against attack by the immune system.
One SOD, which is conserved across the staphylococci, utilizes exclusively manganese to perform this detoxification reaction, whereas the second S. aureus SOD is ‘cambialistic,’ meaning it can function equally well with either a manganese or iron cofactor. This second SOD is unique to the S. aureus group, which are more pathogenic than relatives that lack this metalloenzyme.
Two key amino acids
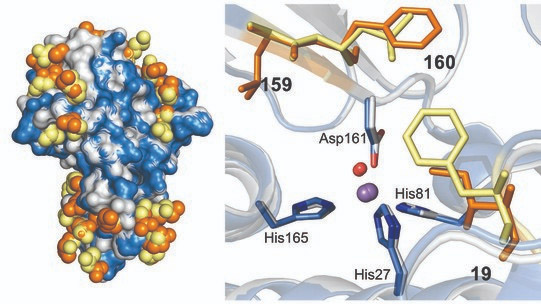
In this study, the biochemical, structural, and biophysical analysis of these SODs with different metal specificity identified two key amino acids in the SOD structure that alter metal specificity. These residues make no direct contacts with the metal-coordinating ligands but control the metal’s redox properties indirectly, demonstrating that subtle architectural changes caused by mutations to amino acids near to the cofactor can dramatically alter metal utilization. A bioinformatic analysis carried out by the team demonstrated a very close evolutionary relationship between these two SODs, suggesting they diverged recently.
“Previous studies suggest that, over time, a metal-dependent protein can switch from one metal to another – an enzyme that uses iron in one organism may have evolved to utilize copper in another. However, ours is the first study to show how evolution can achieve this switch through subtle changes to the enzyme’s structure,” Waldron said.
S. aureus experiences manganese-starvation during infection, implying this may have driven a need for its important manganese-enzyme to switch to using an alternative metal ion when it developed the ability to cause infection.
“The differential importance and close evolutionary relationship between the two staphylococcal SODs, combined with the ability to manipulate the metal they utilize, provided us an opportunity to determine if stresses within the host, such as metal starvation, can drive metalloenzyme evolution,” Kehl-Fie said.
Introducing the mutations identified by the team into living S. aureus cells, which diminish the ability of the cambialistic SOD to use iron, reduced the ability of the bacterium to resist superoxide stress when metal starved by the host.
“This suggests that small changes in metal-dependent activity, in conjunction with stresses encountered within the host, can drive the evolution of metalloenzymes with new cofactor specificity,” Kehl-Fie said.
“Crucially, our analyses have uncovered the mechanism by which evolution has shaped the properties of these metalloenzymes at the molecular level, resulting in a pair of enzymes that utilize different metal ions for catalysis. We propose that this was selected for by the manganese-deficient conditions this bug experiences when it encounters the immune system,” Waldron said.
The study illustrates how evolution has shaped metal utilization by making minor alterations to the chemical environment of the redox-active metal cofactor.
Based on the current investigation the team propose that the shift in metal usage by metalloenzymes of S. aureus has been shaped by changes in the metals available to the bacterium as it evolved from a commensal to an opportunistic pathogen lifestyle.

