Illinois Team Tracks COVID ‘Spike’ Protein for 2020 iGEM Competition
UPDATE: The Illinois team’s “Viralizer” web tool brought home a silver medal at the International Genetically Engineered Machine (iGEM) 2020 competition. Working virtually, the six-member student team created the tool to model the COVID-19 spike protein as it mutates and chart its spread, to aid in the development of new drugs & vaccines.
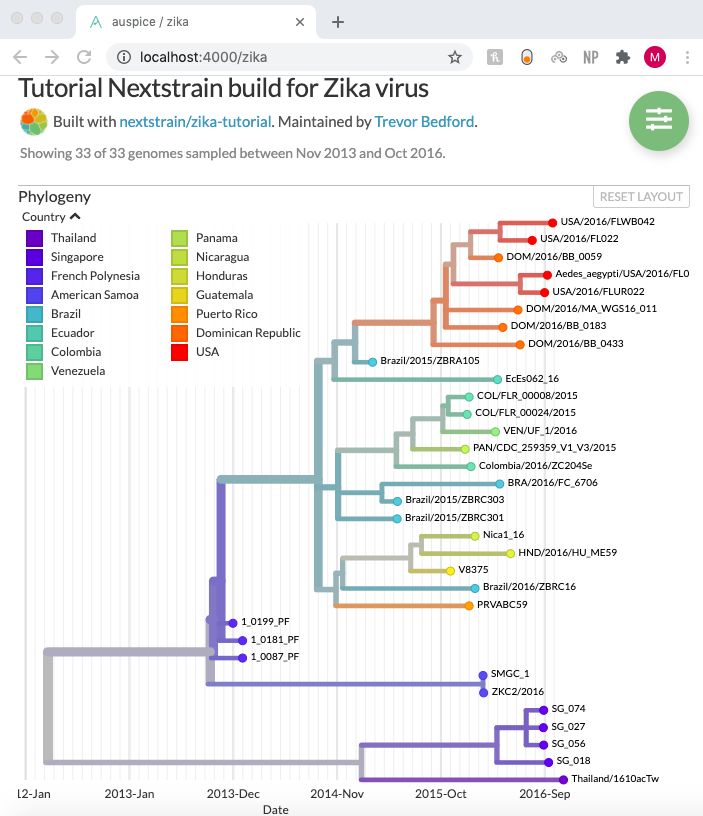
Viralizer details more than 20,000 mutated protein sequences, as well as several hundred potential antibodies designed by the team to neutralize the spike protein. A phylogenetic tree characterizes the progression of the pandemic over space and time. Mary Cook, a junior in bioengineering, said the team was happy with the silver medal and accomplished “everything we hoped for in terms of our antibody design, our phylogenetics output, and the database models uploaded on our website.”
Suva Narayen, a junior in bioengineering who worked on antibody design, said the project turned out better than expected given the amount of work required — and some late nights — as the deadline approached. “Fortunately we were able to finish on time, and the judging and poster sessions went really well for us,” Narayen said.
This year’s virtual Jamboree “definitely felt different” and presented some logistical challenges, Cook said. “However the interactive poster that we put together was a cool way to display our work and we enjoyed still being able to meet all the judges.” The team members are now close friends and hope to continue working together in some way, she said.
“The biggest takeaway from this project for me is the fact that we came up with a solution to an ongoing problem, something I haven’t really done before, Narayan said. Because the COVID-19 pandemic is still being researched, “we had no guidelines to look to if we were stuck. It encouraged me to think outside the box and collaborate with the rest of the team more effectively.”
You can watch the full 18-minute video about the team’s Viralizer project here; read our August article about the project below.
The COVID-19 pandemic created unprecedented challenges for a worldwide competition that brings high school and college students together to tackle big questions in synthetic biology.
But it also provided a unique research opportunity for the University of Illinois team competing in this year’s International Genetically Engineered Machine (iGEM) contest.
The six undergraduates are pooling their talents — remotely — to contribute to the fight against SARS-CoV-2, the coronavirus that causes COVID-19. They’re creating a web tool to build visual models of a key part of the virus as it mutates — specifically the infamous “spike” protein that allows it to attack human cells so easily. The hope is to give researchers crucial information about the virus as they design new drugs and vaccines.
“Our database will be real time, and it will allow researchers to upload their newly collected coronavirus sequences and get those newly mutated protein models for their research,” said Yan Luo, a sophomore in bioengineering on the Illinois team.
Mentored by graduate students and postdocs, iGEM teams meet in the spring to brainstorm ideas for innovative synthetic biology research and develop them over the summer. The projects culminate at the annual Giant Jamboree, iGEM’s international competition, to be held virtually this year in November. The event attracts teams from nearly every continent who can earn bronze, silver, or gold medals by achieving certain standards. Overall winners are chosen from the gold-medal teams.
The Illinois iGEM team is sponsored by the Carl R. Woese Institute for Genomic Biology (IGB) and the Center for Advanced Bioenergy and Bioproducts Innovation (CABBI). Illinois has competed in iGEM since 2008, earning bronze or silver medals and occasionally gold, said Christopher Rao (BSD/CABBI/MME), Professor of Chemical and Biomolecular Engineering (ChBE), who helped launch the first iGEM team and is advising this year’s squad.
Past Illinois projects included a bacterial filing cabinet and a medical biosensor. Last year’s group took an environmental turn: developing a micro-organism to degrade glyphosate, the key ingredient in Round-Up and a suspected carcinogen. Their work showed promise, and with one student returning for this year’s competition Rao hoped the 2020 team might carry on that research.
“Then COVID happened,” said Rao, who also serves as Deputy Leader of CABBI’s Conversion Theme. “There’s no way we could do hands-on training of undergraduates in this environment.”
They decided on a purely computational project, which would be far easier to manage remotely. Given the pandemic, the students wanted to do something COVID-related, Rao said.
“We all just wanted to start fresh and have our own idea,” said Mary Cook, a junior in bioengineering.
Besides Luo and Cook, the team includes Sachin Jajoo, a senior in molecular and cellular biology; Angela Yoon, a junior in integrative biology; Suva Narayan, a junior in bioengineering; and Royal Shrestha, a sophomore in biochemistry. They are mentored by CABBI researchers Matthew Waugh, a chemistry postdoc, and ChBE graduate students William Woodruff and Carl Schultz. CABBI Research Coordinator Anna Fedders and IGB Outreach Activities Coordinator Daniel Ryerson provide administrative support.
The students started work in March, after the coronavirus shutdown. They each came up with an idea for a computational project with synthetic biology applications that would fill a research need. Shrestha suggested a 3D database as an interactive way to show viral structures and help scientists learn more about SARS-CoV-2.
The team presented the idea to their mentors, who helped focus it into a useful deliverable. The students also consulted with professors across campus involved in COVID-related research, including Erik Procko in biochemistry, Mohammed El-Kebir in computer science, and two faculty members in ChBE — Huimin Zhao (BSD leader/CABBI/MMG) and Diwakar Shukla.
Those conversations narrowed the project to spike proteins specifically, Cook said. The team also decided to try, to design an antibody with the information they collected, rather than just a database.
SARS-CoV-2 is mutating — in small ways, but enough to potentially change how its proteins bind to human receptor cells or to a vaccine, Luo said. So researchers want to fully understand the structure of all the mutated proteins, which is a complex process.
One of the most crucial is the spike protein — named for the protrusions it causes on the surface of the coronavirus — which allows the virus to penetrate human host cells and cause infection. Any mutations could alter its infectiousness or impact the efficacy of a future vaccine.
“That is the problem that we are trying to tackle,” said Luo, who is leading the project’s software aspects. “We have thousands of sequences of the spike proteins, but we don’t know their structure. We’re taking the mutated sequences and the already available models of proteins to predict the structure of the mutated spike proteins.”
Some vaccines work through a neutralizing agent that disables the virus, almost like an antivenom, by binding with it and preventing it from interacting with the human body, Luo said. Most researchers design those neutralizing agents in wet labs, testing them hundreds of times until they find the right one. But they usually work with just one strain and don’t necessarily focus on the protein structure.
There’s no current database dedicated to producing a model for the spike protein as it mutates, Luo said. Of the 8,000-plus spike proteins found on the Global Initiative on Sharing Avian Influenza Data (GISAID) platform, “a very, very small percentage have crystal structures that are known through experimentation,” Cook said.
To build their web tool, the students are using NextStrain, an open-source project that pulls together data and visualization tools for viruses and other pathogens to track the spread of outbreaks and improve the public health response. They can upload their own protein sequences, models, and geographic data to make a phylogenic tree showing different spike protein strains in each location.
In the spirit of iGEM, their tool will be open-source. A researcher who discovers a tiny “point mutation” — a change in only one or two nucleotides from the original viral genetic sequence— will be able to enter the new sequence into the tool, which could then predict whether that mutation would make a significant difference in the crystal structure and produce a 3D model of what it would look like.
Most point mutations wouldn’t have a huge effect. But researchers have already found that a variation in one amino acid on the spike protein increased the infectiousness of SARS-COV-2 a thousand-fold, allowing it to spread quickly and become the dominant strain in Europe and the United States. The team hopes to be able to predict which genetic mutations might cause problems in the future.
Cook and Yoon are working on the visualization and website components of the project. Jajoo, an iGEM veteran, is supervising administrative aspects, and Narayan and Shrestha are heading up antibody design.
They are using a popular modeling software called Py Rosetta to compare their models for thousands of spike protein variations with simulated mutations of the human antibody. The software can analyze how they bind together, indicating which antibody offers the best defense and could be developed into a vaccine or drug.
Collaborating remotely has been a challenge. The entire team has never met in person. The online meeting format made it tough initially for students to open up, Rao said, and they didn’t have the usual bonding experience of working together in the lab. While most of the students are in Champaign-Urbana or Chicago, others are several time zones away in California and South Korea. But they’ve all adapted.
“We’ve already had some successes. I think we’re on a good track,” Luo said.
The project has been an enormous learning experience, on many levels. To write the tool, the students had to learn about viruses, programming, bioinformatics, DNA sequences, protein modeling, viral structures, graphic software, and antibody design. They’ve also had to manage expenses, divvy up tasks, and help each other sort through problems.
Connecting with professors and learning about their research was especially valuable, Cook said.
“Most people just don’t realize how much effort scientists are putting in on tackling the coronavirus,” Luo said, from vaccine development to understanding the virus.
Perhaps most important: the ability to design their own project and do research from the ground up. Choosing what problem to work on, and where you can make a difference, is one of the most important — and challenging — aspects of science, Rao said, separating the “true greats” from everyone else.
“Any research that I’d participated in before has been with a graduate student or with a mentor who had the project and the goals laid out,” Cook said. “Here, we had to start from scratch. That has been probably the most rewarding part of iGEM.”

