Comparative Genomics, Illinois
University of Illinois at Urbana-Champaign
Poster Gallery
Comparative Analyses of Villus and Crypt Small Intestinal Cell Gene Expression Profiles
January 10th, 2004
Plant and Animal Genome XII, San Diego, CA. Abstract P835.
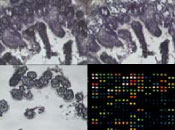 The objective of this study was to compare gene expression profiles of villus and crypt intestinal cell populations within and between species. Laser capture microdissection (LCM) was used to isolate individual villus and crypt epithelial cells from swine, canine, and murine ileal samples. RNA was isolated and amplified using the PicoPure™ RNA Isolation Kit and RiboAmp® RNA Amplification Kit (Arcturus), respectively. Gene expression profiles were generated by hybridizing amplified RNA from a 12 wk-old C57Bl/6 mouse with the NIA 15K Mouse cDNA microarray, amplified RNA from a white crossbred sow with the 13K Porcine Oligo Array (Qiagen) and amplified RNA from a 1 yr-old beagle dog with the Affymetrix Human U133A microarray. Preliminary results show that >1000 genes were more highly expressed in the crypt epithelial cells than in villus cells. This list includes many genes related to apoptosis, cell cycle, DNA replication, and energy/metabolism. Genes (13%) more highly expressed in villus than crypt were associated with matrix or structural proteins. The use of LCM provides a cell-specific gene expression profile of distinct intestinal cell populations. While villus cells are composed of differentiated cell populations possessing specific functions, crypt cells are primarily composed of undifferentiated stem cells. Research in this area may identify factors associated with cellular differentiation and lead to the development of therapies for intestinal disease and may be used as a screening tool for gene targets associated with growth promotion. Supported by Pyxis Genomics, Inc. and the Critical Research Initiative (CRI) at the University of Illinois.
The objective of this study was to compare gene expression profiles of villus and crypt intestinal cell populations within and between species. Laser capture microdissection (LCM) was used to isolate individual villus and crypt epithelial cells from swine, canine, and murine ileal samples. RNA was isolated and amplified using the PicoPure™ RNA Isolation Kit and RiboAmp® RNA Amplification Kit (Arcturus), respectively. Gene expression profiles were generated by hybridizing amplified RNA from a 12 wk-old C57Bl/6 mouse with the NIA 15K Mouse cDNA microarray, amplified RNA from a white crossbred sow with the 13K Porcine Oligo Array (Qiagen) and amplified RNA from a 1 yr-old beagle dog with the Affymetrix Human U133A microarray. Preliminary results show that >1000 genes were more highly expressed in the crypt epithelial cells than in villus cells. This list includes many genes related to apoptosis, cell cycle, DNA replication, and energy/metabolism. Genes (13%) more highly expressed in villus than crypt were associated with matrix or structural proteins. The use of LCM provides a cell-specific gene expression profile of distinct intestinal cell populations. While villus cells are composed of differentiated cell populations possessing specific functions, crypt cells are primarily composed of undifferentiated stem cells. Research in this area may identify factors associated with cellular differentiation and lead to the development of therapies for intestinal disease and may be used as a screening tool for gene targets associated with growth promotion. Supported by Pyxis Genomics, Inc. and the Critical Research Initiative (CRI) at the University of Illinois.