Integration of gene regulatory networks in understanding animal behavior
For years, scientists have attributed animal behavior to the coordinated activities of neuronal cells and its circuits of neurons, known as the neuronal network (NN). However, researchers are pushing the boundaries in understanding animal behavior through the integration of gene regulation.
Fueled by a long-time collaboration with IGB Director and entomology professor Gene Robinson (GNDP), incoming IGB Director of Computational Genomics and computer science professor Saurabh Sinha (BSD/CABBI/GNDP/GSP) helped organize a workshop on “Cis-Regulatory Evolution in Development and Behavior” in 2018 to push a new line of thinking.
“One of the remarkable findings from a study led by Gene and his collaborators was that more eusocial insects seemed to have something different about their regulatory genome,” said Sinha. “It seemed that there was some sort of evolutionary signature of complex social behavior that we hadn't really expected and was one of those findings that really made you re-think the implications.”
The two-day workshop brought together people from a diverse set of skillsets where ideas were exchanged and challenged during discussions on various topics. Two years later, the results of those discussions culminated in a perspective article published in the Proceedings of the National Academy of Science.
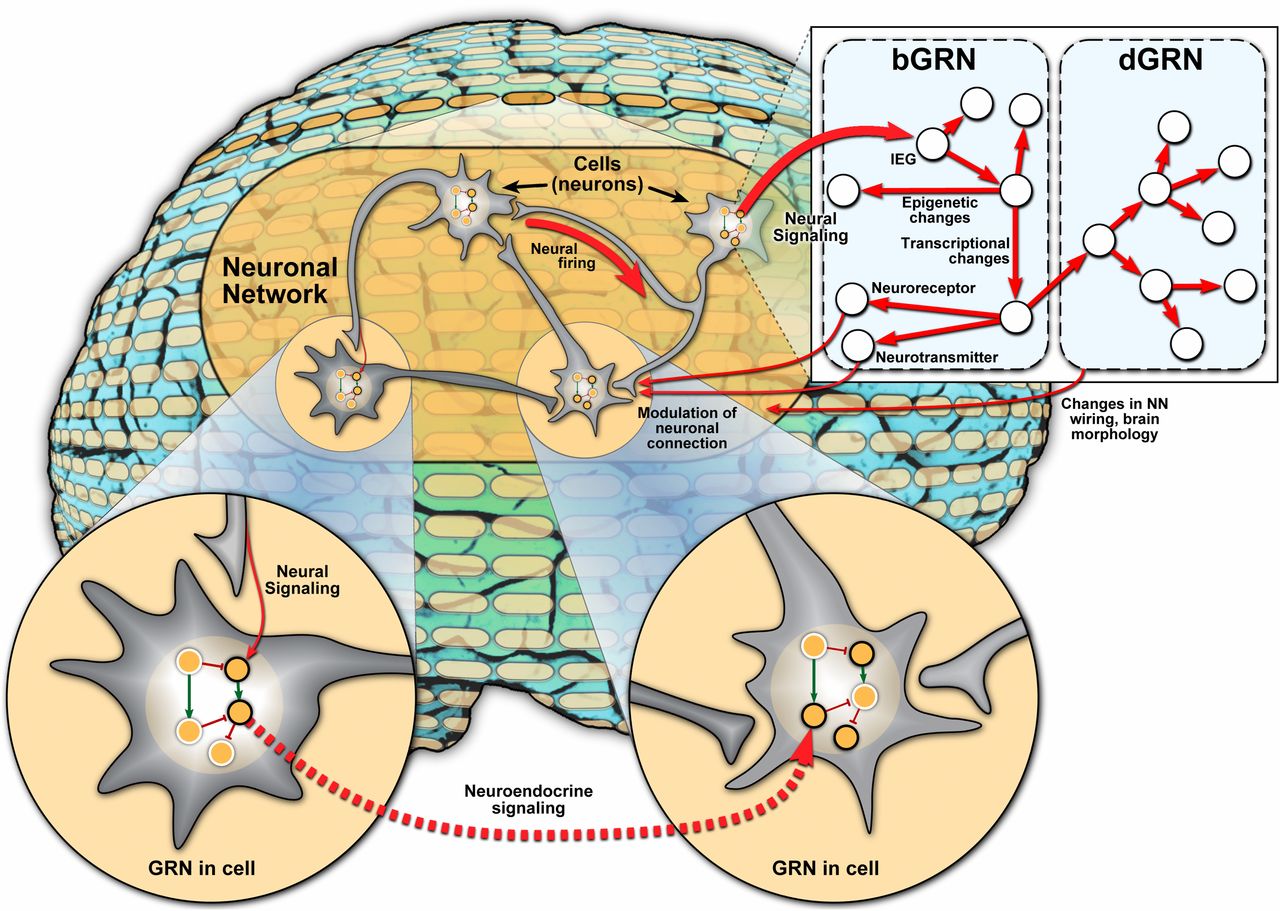
“The starting point for this perspective is that the NN is the de facto standard for understanding what goes on in the brain as pertinent to behavior,” said Sinha. “Our goal was to highlight another level of dynamics that accompany behavior and not just the dynamics of the NN.”
The authors of the perspective synthesized current evidence on the role of the gene regulatory networks (GRNs) – a collection of regulatory interactions between genes – in the context of animal behavior along with the NN. Behavior-associated GRNs (bGRNs) impact gene expression changes associated with a certain animal behavior while developmental GRNs (dGRNs) influence development of new cells and connections in the brain. The integration of NNs, bGRNs and dGRNs across multiple scales holds potential in understanding how these networks work in concert to regulate animal behavior.
“Our first goal was to simply emphasize the significance of the GRN in the behavioral context, before speculating on how the GRN might interact with the NN since current research is lacking,” said Sinha. “One example of an interaction between the NN and GRN could be the modulation of neuronal transmission activity through control of protein or peptide expression by the GRN.”
Through experimental mapping of these networks, the changes in gene expression can be corresponded with behaviors in different cell types. Emerging technologies will play a key role in these efforts. “Measuring gene expression in the brain has been fraught with the heterogeneity of the brain where you have so many different cell types,” said Sinha. “The fact that we have single-cell technology really taking off means that we can have a proper resolution of GRNs in the brain and therefore, examine how cell type-specific GRNs interact with signal transmission through the NN.”
The perspective also touches on how environmental factors and social behavior affect GRNs, which then go on to modulate NN function and behavior. “The environment can induce epigenetic and longer-lasting changes that then lead to the GRN becoming different,” said Sinha. “Looking at brain function not only through the lens of the NN but also through GRNs allows us to bring in the environment in a credible way. In regard to social behavior, there is probably a difference in the GRN of more eusocial bees and that is a starting point for the intriguing possibility that social behavior has some unique characteristics in its GRNs.”
With the emergence of technologies, future analyses of bGRNs and the interchange between bGRNs, dGRNs and NNs in various behavioral contexts will provide a deeper understanding of animal behavior.
