Spinning Disk Demo - February 4th - 27th
Core Facilities at the IGB is your resource for biological microscopy and image analysis. Our facilities are conveniently located, and we provide a variety of services to both IGB researchers and others in the campus community. Our facility is meant to encourage innovation – try new techniques and approaches to achieving your research goals without a significant investment in instrumentation or time.

View Gallery
1
/
10
What’s new at the Core
- High-speed Volume Imaging from Zeiss
-
Zeiss will demo a new Lightfield 4D system in the IGB Core starting November 5th. Matt Curtis will be presenting Lunch with the Core that Wednesday presenting the capabilities of the instrument. The Zeiss Lightfield 4D will collect 80 volumes per second with 2.2micron resolution and fields of view that you would expect from a 10X or 40X objective. This unprecedented combination of speed, resolution, and sensitivity is ideal for time lapse imaging or imaging large sample volumes. More info.
- The LSM 980 is now open for researchers.
-
The LSM 980 is now open for researchers. Please use the training request form to schedule a training. More information about the microscope can be found here.
For questions regarding the LSM 980, please contact Umnia Doha - udoha2@illinois.edu .
- The IGB Core is moving to BookitLab for instrument tracking
-
The IGB Core has purchased BookitLab for tracking instrument use, reserving instrument time and processing billing. The Core plans to setup BookitLab on the LSM 700 April 15th. The April 17th Lunch with the Core will be dedicated to introducing the BookitLab software to existing users. The plan is to transition all instrument tracking to BookitLab by early May. The IGB BookitLab portal can be found here.
Please let us know if you have any questions.
- Midwest Advanced MINFLUX and Super Resolution Workshop 2024
-
Save the Date!
The IGB Core Facilities is hosting the 2nd Annual Midwest Advanced MINFLUX and Super Resolution Workshop from May 20th to 24th. You can find updated information here.
- Lunch with the Core - Spring 2024
-
The IGB Core Facilities is hosting a Spring 2024 seminar series covering the instrumentation in the Core and some of the research taking place in the Core. The seminars are typically held from 12:00 - 1:00 pm in the Carl R. Woese Institute for Genomic Biology, Room 612 (Concourse level). Pizza will be provided to attendees. Please check the Lunch with the Core news posts and weekly emails for updates.
February 14, 2024 "Integrating Live Confocal Microscope Imagery of Plant Stomata with Gas Exchange Measurements" - Joseph Crawford, Postdoctoral Research Associate - Plant Biology
Instrument: LSM 710
Joe's talk can be found here.
February 28, 2024 "Introduction to STEDYCON: A Confocal and STED System with Super-resolution Capabilities" - Umnia Doha, Research Scientist - Core Facilities
Instrument: STEDYCON
Umnia's talk can be found here.
March 6, 2024 "X-Ray Computed Tomography in the Core Facility: Research and Manufacturing Applications of the NSI X5000" - Dr. Austin Cyphersmith, Assistant Director of Research Instrumentation - Core Facilities
Instrument: X5000
Austin's talk can be found here.
March 27, 2024 "3D Data Analysis and Segmentation Tools" - Dr. Kingsley Boateng, Assistant Director of Bioscience Research & Microscopy - Core Facilities
Instrument: Amira & Imaris Software
Kingsley's talk can be found here.
April 3, 2024 "Revolutions in Data" - Dan Davidson, Director of CNRG and Research Computing - Carl R. Woese Institute for Genomic Biology
Dan's talk can be found here.
April 10, 2024 "A solid-liquid superposition model for interfacial solvation structures" - Qian Ai, Graduate Research Assistant - Materials Science & Engineering
Zoom:
https://illinois.zoom.us/j/88982127756?pwd=ME1jZWtZd2txUDBRS3pZa2lUWmNrUT09
Meeting ID: 889 8212 7756
Password: 201556
April 17, 2024 "Introduction to BookitLab" - Dr. Glenn Fried, Director of the Core Facilities
Join Zoom Meeting
https://illinois.zoom.us/j/85934995263?pwd=eE9ZeXNuNFozNCtKdytYMTMyK3dQUT09Meeting ID: 859 3499 5263
Password: 166361April 24, 2024 "Introducing Spatial Transcriptomics to Core Facilities: A Custom Built MERFISH Microscope " - Duncan Nall, Research Scientist - Core Facilities
Instrument: MERFISH
Zoom:
https://illinois.zoom.us/j/87903107373?pwd=R1dYM2dIMmxxTVhWTGNsclNzY2ovdz09
Meeting ID: 879 0310 7373
Password: 783805
May 1, 2024 "Creating Art of Science" - Julia Pollack, Creative Program Manager - Carl R. Woese Institute for Genomic Biology
Zoom:
https://illinois.zoom.us/j/88064323223?pwd=VjJOM1drU2xVOUhHVnM0UUxIL0Q0dz09
Meeting ID: 880 6432 3223
Password: 410927
If you require special accommodations or have dietary restrictions, please email Hannah McClellan - hlm@illinois.edu.
- The STEDYCON has arrived
-
The Core Facilities has purchased a STEDYCON system from Abberior Instruments. This microscope will function as a four color confocal or a two color 2D STED microscope with 40nm resolution. For more information, please visit our STEDYCON instrument page found here.
- Dr. Reza Rajabi-Toustani has joined the IGB Core Facilities
-
Dr. Reza Rajabi-Toustani will be helping with hands-on training with the MINFLUX and other optical microscopes. Previously he was a Postdoc in Joe (Huanyu) Qiao Lab. Reza has been using the MINFLUX from the time it was installed and provided the July 2022 Image of the Month. It can be found here.
- Dr. Umnia Doha has joined the IGB Core Facilities
-
Dr. Doha will be helping with training on the optical microscopes. Umnia comes from the Taher Saif Lab in Mechanical Science and Engineering.
Location
The Core Facilities is located on the concourse level (basement) of the Carl R. Woese Institute for Genomic Biology at the University of Illinois Urbana-Champaign. The entrance is adjacent to Array Café.
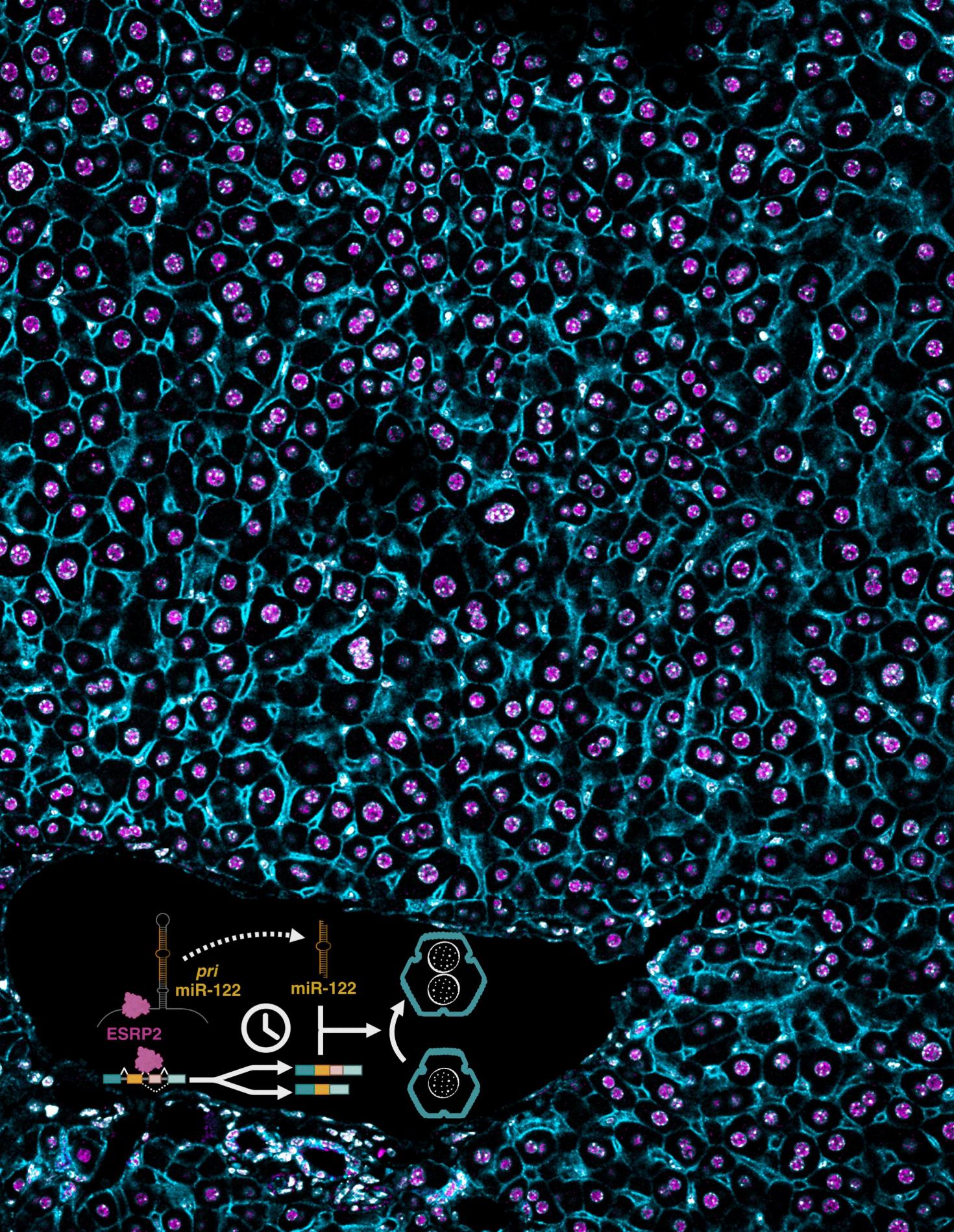
A section of mouse liver stained for ESRP2 and membrane-specific (CTNNB1) marker in a wildtype mouse. Inset: Timed activation of the ESRP2–miR-122 axis promotes hepatocyte binucleation and is linked with promoting hepatocyte quiescence.
Data taken on the LSM 900. Image provided by the Kalsotra Lab associated with this paper https://doi.org/10.1101/gad.352129.124


This image shows the internal construction of the "Motive nozzle", a crucial component of the two-phase ejector, which is widely used in refrigeration and air conditioning applications. This motive nozzle is typically a converging-diverging nozzle through which a high-pressure fluid expands, and upon expansion, a very high velocity (often sonic or supersonic speed) is achieved at the motive nozzle outlet. This part, along with other ejector components, is used to improve system performance in HVAC&R systems. This image was produced using the "North Star Imaging X5000 2D Digital Radiography and 3D Computed Tomography", and implementing the vortex scan technique. This image shows the exterior (left) and interior (right) detail geometry for the motive nozzle, which would provide valuable insights into the dimensional variations for manufacturing these components and how these dimensional variations influence the HVAC&R system performance.
Author: Md Muntasir Alam - Energy Transport Research Lab, MechSE
"Representative coronal image from one hemisphere of an E16.5 mouse embryo brain section (25 μm thick) stained for NeuN (red) and Hoechst (blue). NeuN is a neuronal nuclear protein that serves as a marker of post-mitotic neurons, allowing identification of maturing neuronal populations. Hoechst is a fluorescent DNA-binding dye that labels all cell nuclei, providing structural context. Image was acquired using a Zeiss LSM 880 confocal microscope with a 20× objective. We thank Fernando Rigal for his assistance with microscope settings"
Image provided by Rafael J. Gonzalez-Ricon from the Antonson Lab


While the mechanisms driving activation of the innate immune response during viral infection are well understood, the spatial organization remains poorly defined. Using in vitro models of the respiratory tract, we aim to dissect how spatial structure shapes the activation of the innate immune response during influenza A virus infection.
Axioscan image of human bronchial epithelial cells (HBECs). Stained with DAPI (blue) and Phalloidin (green)
Joel Rivera Cardona from the Brooke Lab


This is an image of a gelatin methacrylate model for the perivascular niche of the bone marrow. The perivascular niche, defined by proximity to arteriolar and sinusoidal vessels that comprise endothelial and mesenchymal cells within the bone marrow, regulates hematopoietic stem cell proliferation, differentiation, and quiescence through secreted factors, cell-cell interactions, and local remodeling of the bone marrow.
In this image, we have Hoechst 33342-stained nuclei in blue, murine bone marrow-derived mesenchymal stem cells (mMSCs) stained for podoplanin in cyan, and murine bone marrow-derived endothelial cells (mBMECs) stained for CD31 in green.
Gunnar Thompson from the Harley Lab
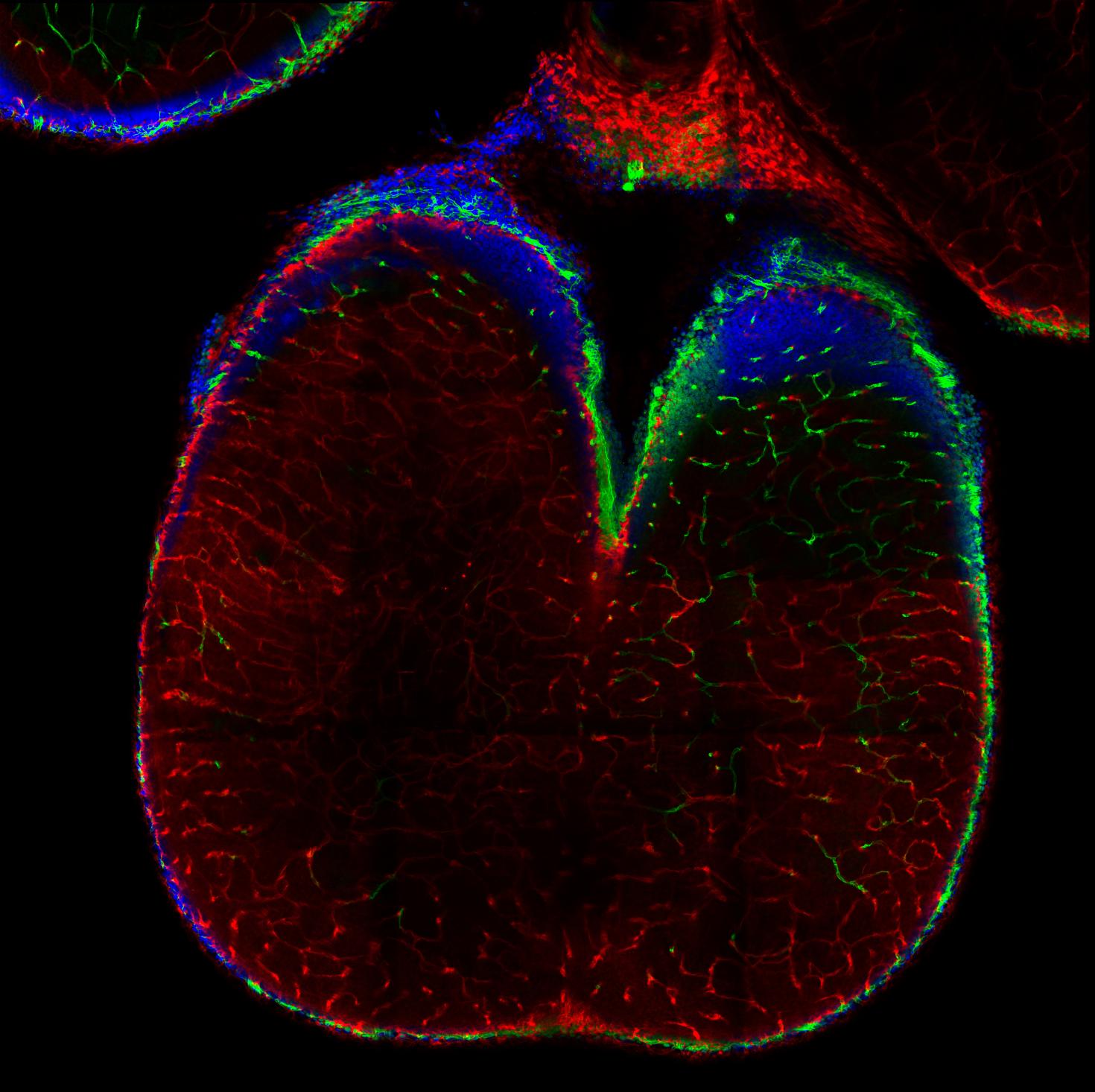

Two-photon image deep into the whole brain of a mouse fetus (Gestational Day 16.5) and post-processed with the Spectral Unmixing Module. Shown is the cerebellum structure and labels are for: CD31 (Green), Claudin-5 (red), Hoechst (Blue). The fetus was cleared with an adapted form of a new tissue clearing technique called ADAPT-3D.
Image taken with LSM 980
Image Courtesy: Fernando Rigal from the Antonson Lab


3D reconstruction of Salmonella Typhimurium biofilms in presence of differentially charged micro-nanoplastics (MNP). The study examined how MNP exposure influences bacterial attachment, aggregation, and extracellular polymeric substance (EPS) production- key factors that enhance biofilm resilience in food processing environments. Confocal laser scanning microscopy (CLSM) was used to visualize biofilm architecture and development over time. Understanding these interactions is critical, as enhanced biofilm formation may increase bacterial persistence, antimicrobial resistance, and the risk of contamination, posing serious threats to public health.
LSM 880, stained with FilmTracerFM 1-43 green biofilm cell stain
From Jayita De in the Pratik Banerjee Lab


An unexpected discovery of a bird skeleton within a nest built in a wooden enclosure provides new insights into nesting behavior and material composition. This finding informs the ongoing design of a concrete nest, exploring the intersection of natural instincts and architectural innovation.
Author: Arpit Amarseda

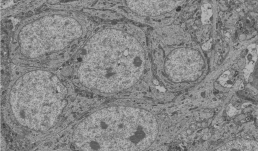
"This SBF-SEM image displays the cell body of a CA1 pyramidal neuron in the hippocampus. Our study aims to investigate mitochondrial morphology and complexity in the mouse hippocampal neurons."
Image taken on Zeiss Sigma VP SBF-SEM
Authors: Kingsley Boateng (IGB Core) and Vipendra Kumar (Nien-Pei Tsai’s
Lab).

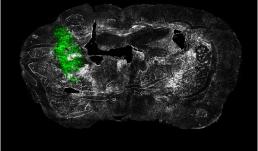
"Ex vivo coronal section of glioma cells, labeled with GFP (green), in a mouse model of glioblastoma (GBM) on day 14 of tumor development."
Image taken on Axioscan.
Author: Urbi Saha - Andrew Smith's Lab


