Researchers discover a starring role for chaperone protein Hfq in gene regulation
A cell’s efforts to respond and adapt to its external environment rely on an elaborate yet coordinated set of molecular partnerships within. The more we learn about this complicated internal dance, the more we appreciate the flexibility of its roles. In a recent University of Illinois study, graduate student Muhammad Azam and Professor of Microbiology Cari Vanderpool have demonstrated that a protein typically assumed to support the functions of other molecules is actually able to assume a primary role itself.
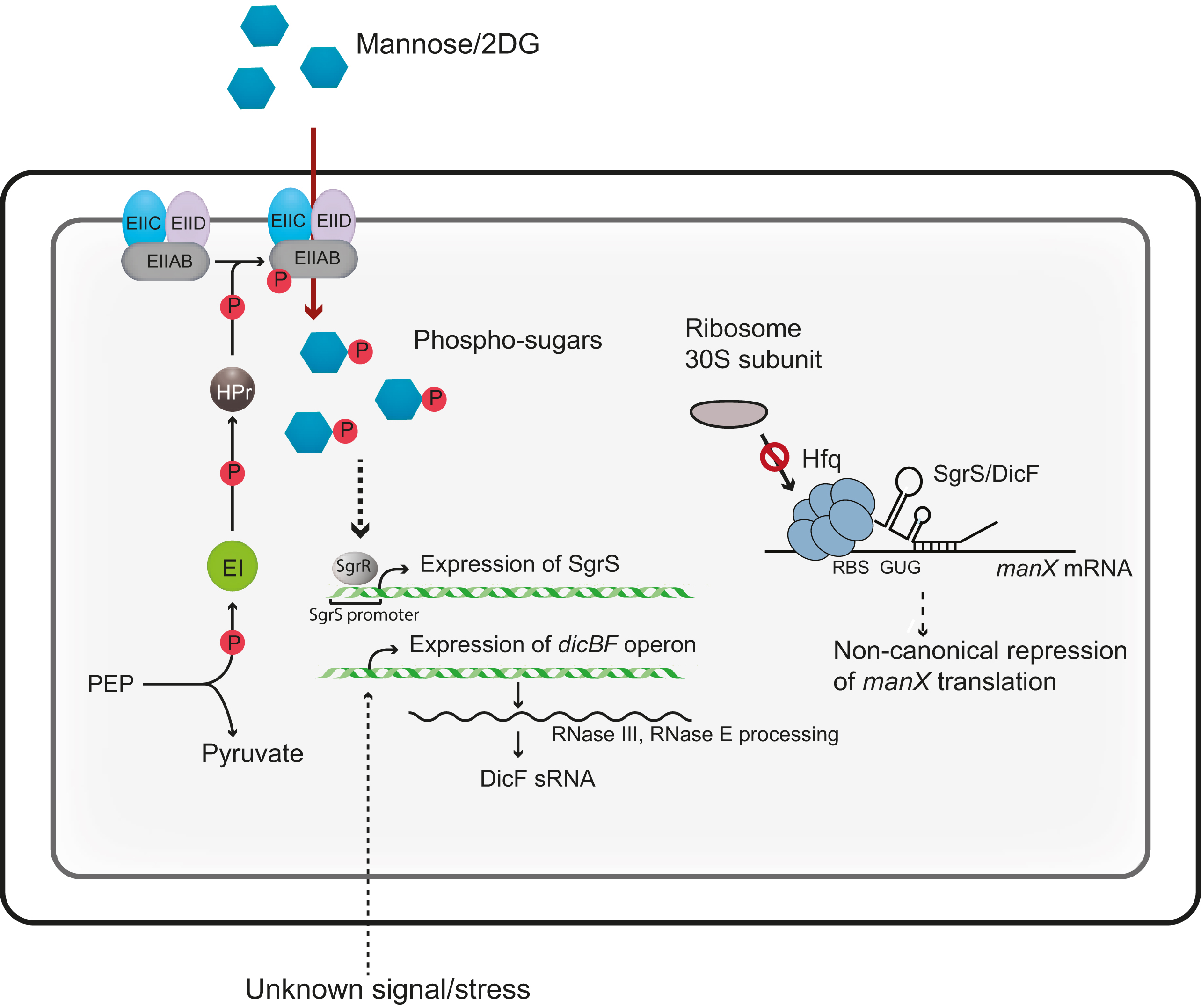
The work, published in Nucleic Acids Research and supported by the NIH, is part of a long-term effort in the Vanderpool laboratory to understand how bacterial cells balance uptake of sugars from their surroundings with their metabolic needs. Vanderpool is also a member of the Carl R. Woese Institute for Genomic Biology, participating in the thematic research group Microbiome Metabolic Engineering, which explores the relationships between microbial communities and the environment.
“For a long time, we've been trying to understand how bacterial cells coordinate uptake of sugars with their metabolism because if these processes become unbalanced, cells get sick and stop growing,” Vanderpool said. “We study a genetic regulatory system in E. coli and related bacteria that help them keep this balance between sugar uptake and metabolism.”
One way that bacteria adapt to environmental change is by adjusting gene activity. Many genes are like recipes for proteins; machinery in the cell reads the information captured in the DNA sequence of a gene and produces a complementary strand of messenger RNA (mRNA). This information enables another piece of molecular machinery called a ribosome to construct the corresponding protein. One way bacteria can adjust the amount of protein produced is by increasing or decreasing the number of times ribosomes “read” the relevant mRNA.
For example, bacteria possess multiple genes coding for proteins that transport sugars into the cell. In an environment with an oversupply of sugar available, specialized molecules may bind to the mRNAs that produce those proteins and prevent ribosomes from reading them as easily, resulting in fewer transporters and a slower uptake of sugar. Vanderpool and her colleagues are interested in the details of how that regulation occurs.
“If we understand this process better, we might eventually be able to devise new strategies to manipulate growth of bacteria that live in us and on us—improving the growth of helpful bacteria or inhibiting the growth of harmful bacteria,” she said.
In Azam and Vanderpool’s study, the focus was the regulation of a gene called manX that codes for a protein able to transport mannose, a sugar, into the cell. They were particularly interested in how a molecule called SgrS helps control how much ManX protein is made.
SgrS is actually a short strand of RNA, the same type of molecule used to transfer the message of a gene to ribosomes during protein production. Small RNAs (sRNAs) like SgrS can play a regulatory role in this process by sticking to the part of an mRNA where a ribosome usually begins its work. Some sRNAs prevent the ribosome from grabbing the mRNA, while others help it hang on, respectively decreasing or increasing the amount of protein made.
“Prior to this study, we knew that SgrS helps cells by slowing down sugar uptake to allow metabolism to catch up,” Vanderpool said. She and Azam began to examine how SgrS does this, and how it might be supported in this function by a protein called Hfq. Hfq is an RNA chaperone protein, a type of protein whose name evokes its function; Hfq helps guide and stabilize sRNAs as they bind to mRNAs.
Azam and Vanderpool were surprised to discover that SgrS does not appear to cling to the part of the manX mRNA where a ribosome needs to attach. Instead, the Hfq protein grabs onto this area, while SgrS pairs up with a site further along the mRNA strand, reversing the roles that researchers have consistently observed in bacterial gene regulation.
“Hfq, not the sRNA, is the direct repressor,” Azam said, describing the mechanism that the study revealed. “This is an unusual mode of repression . . . SgrS acts as a guide to recruit Hfq” rather than Hfq helping to guide and stabilize SgrS.
“I was very excited that we were able to define a specific mechanism for this regulation,” Vanderpool said. “Our work is the first to have shown a specific and new mechanism by which small RNAs and Hfq cooperate to control translation of mRNAs.”
Vanderpool and Azam suggest that this discovery of a reversed relationship between a sRNA and Hfq may lead to further documentation of similar mechanisms in bacteria—researchers may have missed other examples in the past because they didn’t know to look for them. They hope that a more comprehensive understanding of gene regulation in bacteria will eventually enable scientists to reprogram bacterial behavior by affecting gene activity.
