Researchers use computation and genomics to battle tooth decay
An expert in using computational and experimental techniques to combat infectious diseases, Illinois Bioengineering faculty and MMG member Paul Jensen is taking aim at one of the most prevalent chronic diseases in the U.S.—tooth decay. Earlier this year Jensen received $218,000 in funding from the National Institute of Dental and Craniofacial Research (NIDCR) to apply a data-driven approach to understanding the role that certain bacteria play in cavity formation.
For years, scientists have known that cavities occur when the good and bad bacteria in our mouth become imbalanced. The bad bacteria, Streptococcus mutans, forms a biofilm (aka tartar), then takes the sugars we eat and ferments them into acid, which decalcifies our teeth and causes decay.
According to Jensen, there’s a second deleterious bacteria called S. sobrinus that accelerates tooth decay in some people; however very little is known about this microbe and its function because it’s much more difficult to work with in the lab than S. mutans.
“Although rarer than S. mutans, S. sobrinus makes acid more quickly,” noted Jensen, a research assistant professor based at the Carl R. Woese Institute for Genomic Biology on campus. “If S. sobrinus is present, you’ll have rampant tooth decay.”
With the NIDCR funding, Jensen and his students were the first researchers worldwide to complete the entire genome of S. sobrinus —the S. mutans’ genome was sequenced years earlier. Jensen’s team has also built a genome-scale model of S. mutans, which enables them to study a full network of its metabolism. In all, they have identified nearly 480 different genes and 600 chemical reactions in the network. They are now applying this knowledge to develop a similar model for S. sobrinus.
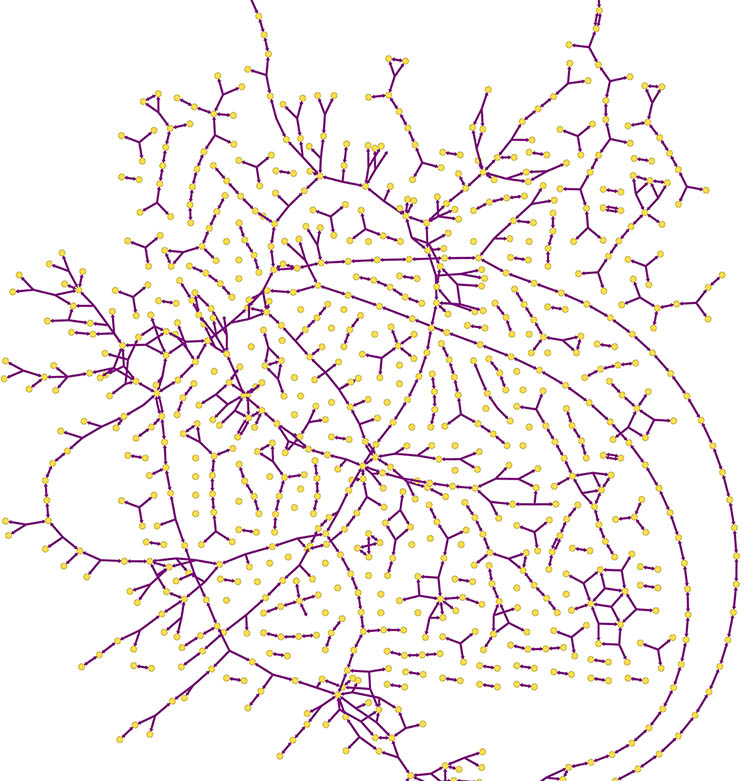
“Once we have a genome-scale model of S. sobrinus, we will be able to computationally do any experiment we want,” said Jensen. “We can simulate what will happen when we delete certain genes or when we feed the bacteria different sugars. We can also put the S. sobrinus and S. mutans models together and see how they interact.”
Ultimately, Jensen aims to use the simulations to develop therapeutic measures that can alter the levels of S. sobrinus and S. mutans in the mouth. For example, his team will create a short list of promising drug targets for further experiments.
“We know these two bacteria make the environment in the mouth incredibly acidic, which involves large numbers of genes in order for them to survive all that acid,” said Jensen, noting that their research could identify places where genes could be targeted in order to change the microbes’ destructive function.
“A lot of scientists are interested in S. sobrinus and they want to see what we find out,” said Jensen. “Using computation is allowing us to rationally demonstrate that this is the way to study S. sobrinus even though other lab-based experiments failed in the past.”
