Group develops gene circuit design strategy to advance synthetic biology
Over the last 17 years, scientists and engineers have developed synthetic gene circuits that can program the functionality, performance, and behavior of living cells. Analogous to integrated circuits that underlie myriad electronic products, engineered gene circuits can be used to generate defined dynamics, rewire endogenous networks, sense environmental stimuli, and produce valuable biomolecules.
These gene circuits hold great promise in medical and biotechnological applications, such as combating super bugs, producing advanced biofuels, and manufacturing functional materials.
To date, most circuits are constructed through a trial-and-error manner, which relies heavily on a designer’s intuition and is often inefficient, said University of Illinois Bioengineering Associate Professor Ting Lu. “With the increase of circuit complexity, the lack of predictive design guidelines has become a major challenge in realizing the potential of synthetic biology,” said Lu, who also is affiliated with the Carl R. Woese Institute for Genomic Biology and Physics Department at Illinois.
Researchers have turned to quantitative modeling to address this gene circuit design challenge. Typical models regard gene circuits as isolated entities that do not interact with their hosts and focus only on the biochemical processes within the circuits, noted Lu.
“Although highly valuable, the current modeling paradigm is often incapable of quantitatively, or even qualitatively sometimes, describing circuit behaviors,” he said. “Increasing experimental evidences have suggested that circuits and their biological host are intimately connected and their coupling can impact circuit behaviors significantly.”
Lu and his graduate students, Chen Liao and Andrew Blanchard, recently addressed the challenge by constructing an integrated modeling framework for quantitatively describing and predicting gene circuit behaviors. Using Escherichia coli (E. coli) as a model host, the framework consists of a coarse-grained but mechanistic description of host physiology that involves dynamic resource partitioning, multi-layered circuit-host coupling, and a detailed kinetic module of exogenous circuits.
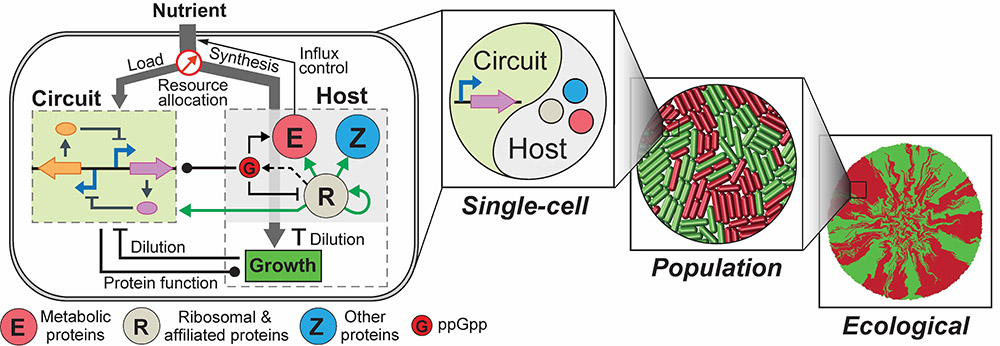
The team demonstrated that, following training, the framework was able to capture and predict a large set of experimental data concerning the host and simple gene overexpression. For instance, they discovered that ppGpp-mediated effects are the key to understanding constitutive gene expression under environmental changes, including both nutrient and antibiotic changes. The team also demonstrated the utility of the platform by applying it to examine a growth-modulating feedback circuit whose dynamics is qualitatively altered by circuit-host couplings and revealing the behaviors of a toggle switch across scales from single-cell dynamics to population structure and to spatial ecology.
Although Lu’s framework was established using E. coli as the model host, it has the potential to be generalized for describing multiple host organisms. “For example, we found that, by varying only a single parameter, the framework successfully predicted several key host metrics, including RNA-to-protein ratio, RNA contents per cell, and mean peptide elongation rate, for Salmonella typhimurium and Streptomyces coelicolor,” said Lu.
According to Lu, this work advances the quantitative understanding of gene circuit behaviors, and facilitates the transformation of gene network design from trial-and-error construction to rational forward engineering. By systematically illustrating key cellular processes and multilayered circuit-host interactions, it further sheds light on quantitative biology towards a better understanding of complex bacterial physiology.
The work is described in Lu’s paper, “An integrative circuit-host modeling framework for predicting synthetic gene network behaviors,” published in the September 25, 2017 issue of Nature Microbiology.
This research was supported by grants from the National Science Foundation, the Office of Naval Research, and the American Heart Association.
