Spatial Omics
Contact us at spatial@igb.illinois.edu.
Spatial omics is an approach to biology research that combines
- genome-scale omics data (gene transcripts, epigenetic markers, proteins, metabolites, etc.) with
- high-resolution spatial information
to better understand molecular, cellular, and tissue-level processes.
- Overview
-
Spatial omics has enormous potential because it connects the rich but abstract world of high-dimensional omics measurements to the three-dimensional, tangible world of physical processes. Biological function is carried out by molecules but fundamentally situated in space, so measuring both these quantities together can give a more complete understanding of how things work. Nature Methods named spatial transcriptomics Method of Year in 2020 and the technology is at the forefront of a new generation of biological research.
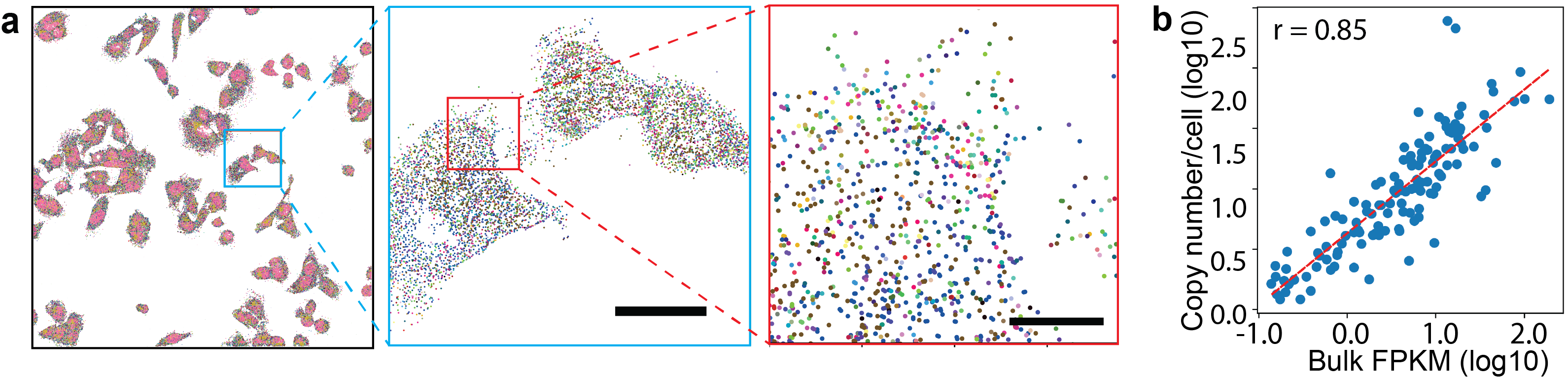
Spatially resolved, single-molecule resolution gene expression in human U2OS cells, imaged by the Han Lab at the University of Illinois. a) Each dot is an RNA molecule and each color represents a gene. b) Gene expression quantified from the spatial omics data are highly correlated with bulk RNA-seq on a biological replicate. This new field is reaching a critical point. Its promise is evident, but its foundational concepts and techniques have not yet been fully codified. There are currently dozens of competing experimental approaches, hundreds of available analytic tools, and likely thousands of researchers formulating the right questions to ask. Spatial omics needs new, paradigm-shifting ideas.
These breakthroughs will likely arise from a convergence of disciplines, because spatial omics pertains to a wide range of areas:
1. its questions can come from biology and medicine,
2. its concepts might borrow from spatial ecology and geographic information systems,
3. its measurement technology requires engineering and instrumentation, and
4. its data analysis needs machine learning and artificial intelligence.Because it crosses disciplines, research in spatial omics promises to catalyze new discoveries across multiple fields. Questions in biology can motivate new directions in engineering and analytics, which in turn can inspire new biological questions.
The strength and diversity of research at the University of Illinois make it uniquely positioned to make major, fundamental contributions to this fast-growing field. The IGB held a spatial omics workshop in March 2022 that was attended by faculty and staff from over eight different departments across campus. The purpose of this Spatial Omics Initiative is to build cross-campus collaborations to continue this momentum.
- Resources
-
Introductory material (Illinois login may be required)
- At our opening workshop, the Han Lab at the University of Illinois presented slides introducing the existing major classes of spatial transcriptomics technology.
- The Han lab has also prepared additional slides introducing one particular type of imaging-based technology.
- See here for a very nice non-technical article about the state of the field.
Review papers
The following papers provide a good introduction to the field, targeted to a biological audience.
- Bressan, D., Battistoni, G., Hannon, G.J. "The dawn of spatial omics". Science 381, 2023, eabq4964. https://doi.org/10.1126/science.abq4964
- Moffitt, Jeffrey R., Emma Lundberg, and Holger Heyn. “The Emerging Landscape of Spatial Profiling Technologies.” Nature Reviews Genetics, July 20, 2022, 1–19. https://doi.org/10.1038/s41576-022-00515-3.
- Moses, Lambda, and Lior Pachter. “Museum of Spatial Transcriptomics.” Nature Methods 19, no. 5 (May 2022): 534–46. https://doi.org/10.1038/s41592-022-01409-2.
- Palla, Giovanni, David S. Fischer, Aviv Regev, and Fabian J. Theis. “Spatial Components of Molecular Tissue Biology.” Nature Biotechnology 40, no. 3 (March 2022): 308–18. https://doi.org/10.1038/s41587-021-01182-1.
- Rao, Anjali, Dalia Barkley, Gustavo S. França, and Itai Yanai. “Exploring Tissue Architecture Using Spatial Transcriptomics.” Nature 596, no. 7871 (August 2021): 211–20. https://doi.org/10.1038/s41586-021-03634-9.
- Tian, Luyi, Fei Chen, and Evan Z. Macosko. “The Expanding Vistas of Spatial Transcriptomics.” Nature Biotechnology, October 3, 2022, 1–10. https://doi.org/10.1038/s41587-022-01448-2.
Analysis tutorials
These introductory tutorials for analyzing spatial transcriptomics data use popular analysis packages and give a good idea of the possible data structures.
- Analysis of grid-based data using the R package Seurat.
- Analysis of grid- and imaging-based data using the Python package squidpy.
- Analysis of imaging-based data using the Python package starfish.
Public datasets
These datasets may be useful for testing out new analytical methods.
Description Link Source Spatial Omics Database Data Yuan et al. (2023) Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex Data Maynard et al. (2021) MERFISH measurements from the mouse hypothalamic preoptic region Data Moffitt et al. (2018) MERFISH measurements in the mouse ileum Data Petukhov et al. (2021) Slide-seq data from mouse hippocampus Data Rodruiques, Stickels et al. (2019) - Working Group
-
The Spatial Omics Initiative organizes a monthly working group. The goal is to build relationships and develop collaborative proposals. Meetings will be held jointly with the IGB Center for Artificial Intelligence and Modeling (CAIM), to develop new ideas at the intersection of biology and analytics. We encourage trainees from both biological and analytical research groups to attend these seminars, to present the work done in their labs, and to participate in collective discussions; this will be a fantastic opportunity to learn how to work with a diverse group of collaborators.
Spring 2024
Every other Monday from 12-2 pm in IGB 2410, from January 22 to May 12, 2024, lunch provided.
This semester the working group will also be offered as a graduate seminar, please register for IB 546, CRN 62171. Students attending for credit are expected to attend every meeting and to present once during the semester. Students are encouraged to present in groups or to share a slot. This is a great opportunity to get feedback and to establish collaborations with other labs.
Please sign up below if you are interested in speaking about:
- Questions about data analysis
- Presentations of new results + feedback
- Presentations of a paper with new techniques/methods
- Practice presentations for conferences/talks
- Feedback on ideas for grant proposals/planned experiments
https://docs.google.com/document/d/1XoDeGKGhc2IvZyILAEYfminfHqS_pawh1xVpp6c30VQ/edit?usp=sharing
We will also invite presentations from people outside the immediate field but who have relevant expertise, for example in artificial intelligence, analysis of spatial data, etc.
Past working group meetings
Fall 2023
September 5, 2023 (Tuesday) on Zoom: Introduction (slides, recording)
October 2, 2023: Proposal discussions
November 6, 2023 hybrid meeting: Adam Stewart on deep learning in spatial data analysis
December 4, 2023: Xin Li on spatial and temporal patterns in neuronal diversitySpring 2023
January 30, 2023: Hee Sun Han and Jane Zhao
February 27, 2023: Andrew Steelman and Yuxiong Wang
April 10, 2023: Auinash Kalsotra and Fan Lam
May 8, 2023: Zeynep Madak-Erdogan and Aiman Soliman